User instruction
Overview
The WDDD provides a search for quantitative information about cell division dynamics and 4D DIC microscope images of wild-type and RNAi embryos.Search
To perform a search for the information and the microscope images:- Select orf, gene or id.
- In the query text box:
A) For orf: Enter the name of an ORF of interest.
B) For gene: Enter the name of a gene of interest.
C) For id: Enter the accession number of the database.
- Press search button, and wait a moment for the results to appear.
The result table provides a list of the information in the database. Each row in the table corresponds to an RNAi embryo for which the information and microscope images were obtained. Columns represent the accession number of the data, orf name, gene name, chromosome number, the information about the corresponding RNAi experiment, the information about the settings of microscope and software, the result of hatch check of the embryo on the glass slides recorded by 4D DIC microscope, and the links to view of 4D DIC microscope images and the information about cell division dynamics, a compressed file for quantitative data, and that for 4D DIC microscope images.

4D view
To view 4D DIC microscope images and the information about cell division dynamics, please press the view button in the "4D view" column in the table. Left panel and right panel represent 4D DIC microscope image and the information about cell division dynamics, respectively. Focal plane can be changed by up or down key, and time point can be changed by left or right key. View angle can be changed by mouse drag-and-move.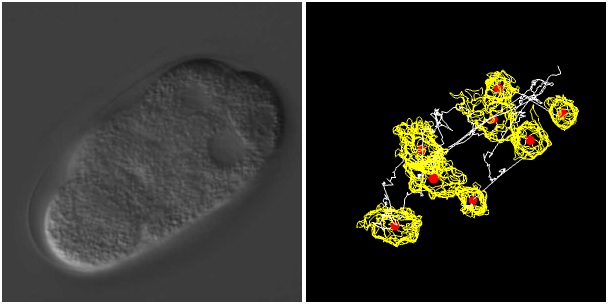
Note:
- This page is programmed by HTML5 with WebGL technology and is designed for the latest version of FireFox web browser. Please use the latest version of FireFox web browser under Windows vista or later, or Mac OS Snowleopard or later.
- In the viewer program, we used application cache technique. Please accept use of offline files cache through the message from browser and wait a minute for the download completion if you want to view microscope images smoothly and view the images and quantitative data synchronously. When a behavior of the viewer is unstable, please clear the offline files cache in your web browser.
CDD data
Quantitative data is provided as a compressed file. This file contains 5 files and a set of image files (*normal quality jpeg files). Two files contain data of quantitative information about cell division dynamics: the ROI file contains the outlines of detected nuclear regions (the file extension is "roi"), and the LNG file contains their transition (the file extension is "lng"). The CRD file contains the information about the embryonic center and the anterior-posterior axis calculated from the embryonic regions (the file extension is "crd"). The other two files are used for viewing software: the IV file, used for the "4D Image Viewer" (the file extesion is "iv"), and the DDV file, used for the "Developmental Dynamics Viewer" (the file extension is "ddv"). All files are text files that can be read by a text editor. The "img" folder contains a set of image files for the viewer.Data format
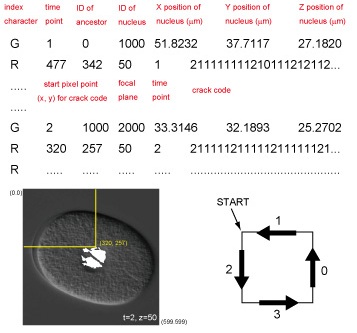
The LNG file contains mainly information about the dynamics of nuclear regions. Each line represents each nucleus at a single time point. The first three numbers of each line represent time point, identification number of the ancestor of the nucleus at the previous time point, and unique identification number of the nucleus. The next three numbers represent the x, y, and z positions of the centroid of the nucleus relative to the upper-left corner of the image and the top focal plane, respectively, where x and y positions are represented by converting 1 pixel to 0.105um. The last string represents the cell name.

IMG data
4D DIC microscope images are provided as a compressed file. The file format of these images is JPEG at a very high quality (* high quality jpeg files). If you develop new quantitative morphological measures, please use these jpeg files that can be downloaded from the DL button in the "img data" column in the result table.Softwares
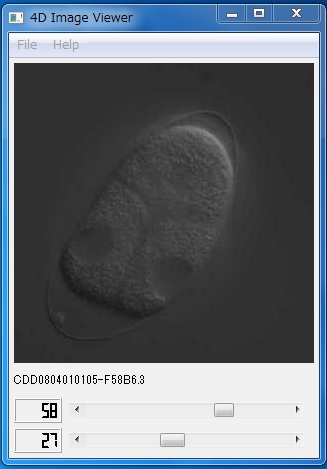
To view 4D DIC microscope images and quantitative information about cell division dynamics, we prepared two desktop application packages, "4D Image Viewer" and "Developmental Dynamics Viewer". These packages run on the Windows platform on a typical IBM-compatible personal computer. The programs were written in C++. Nokia Qt framework was used for the implementation of graphical user interface.4D Image Viewer
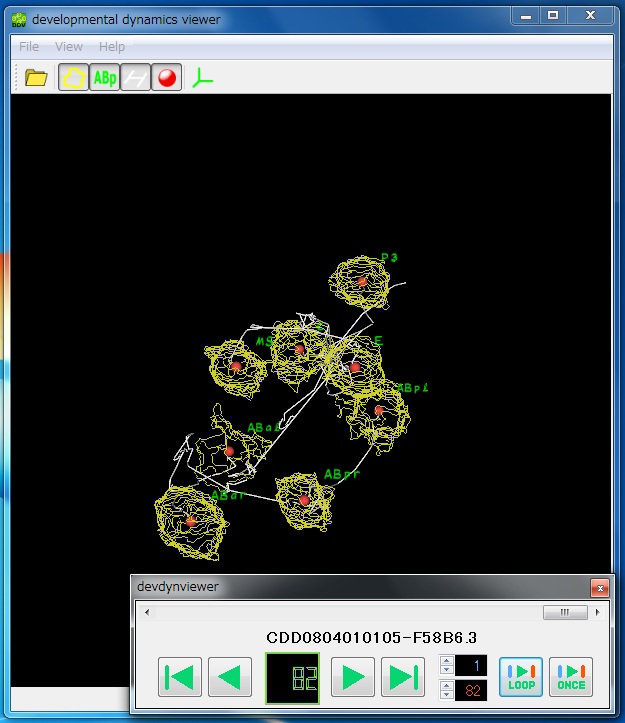
Developmental Dynamics Viewer